
The Swine Health Information Center, launched in 2015 with Pork Checkoff funding, protects and enhances the health of the US swine herd by minimizing the impact of emerging disease threats through preparedness, coordinated communications, global disease monitoring, analysis of swine health data, and targeted research investments.
Set for April 25
The Swine Health Information Center in collaboration with the American Association of Swine Veterinarians will host a webinar titled FMDV Incursions in EU: Situation Update and Considerations for US Prevention. The webinar will be held on Friday, April 25, 2025, from 10:00 am to 11:30 am CST.
Foot-and-mouth disease virus has recently been identified in three European countries after maintaining decades of negative status and poses an emerging disease threat for swine production. The goal of the webinar is to provide the latest information on FMDV including an overview of the virus, clinical signs, global circulation, risks to swine, control and mitigation efforts. Further, the webinar will provide key links to available resources in support of efforts for prevention and preparedness for FMDV and other foreign and emerging diseases.
Current confirmed presenters include:
Jonathan Arzt, DVM, MPVM, PhD, Research Veterinary Medical Officer, United States Department of Agriculture ARS
Overview of FMDV including clinical signs, diagnosis and impact
Maria Sol Perez Aguirreburualde, DVM, PhD, Deputy Director & International Research Development Manager, University of Minnesota
Global FMDV circulation
Patrick Webb, DVM, Assistant Chief Veterinarian, National Pork Board
Available resources for FMDV prevention and preparedness efforts
Gyula Balka, DVM, PhD, Associate professor Department of Pathology
University of Veterinary Medicine, Budapest, Hungary
Denise Wuellner, DVM
PIC Deutschland, Health Assurance, Central and Eastern Europe
Drs. Balka and Wuellner will discuss the EU perspective on identification, management, and control of FMDV.
This webinar, hosted by SHIC and the American Association of Swine Veterinarians, is conducted by the Swine Medicine Education Center at Iowa State University.

The Swine Health Information Center funded a study to evaluate tongue fluids (TF) samples from stillborn pigs as an indicator of PRRS detection in liveborn littermates. Led by Drs. Isadora Machado and Daniel Linhares of Iowa State University, the study investigated TF as a risk-based sample type in commercial breeding herds to predict PRRSV detection in the litter. Litters with stillborn piglets and small litter size had a higher probability of being PRRSV positive. Moreover, TF from stillborn piglets were shown to be a reliable indicator of PRRSV status for their liveborn littermates, indicating that TF can be an effective risk-based approach for PRRSV detection.
Find the study industry summary for project #23-076 on this page.
A risk-based approach to animal selection for sampling enhances PRRSV detection by increasing the probability of selecting an animal harboring the pathogen while requiring a smaller sample size. Aggregated TF from dead animals has emerged as a promising risk-based approach. It has a similar PRRSV RNA positivity rate as serum, processing fluids, and family oral fluids. Its composition is complex, especially in stillborns, consisting of blood, saliva, respiratory mucus, amniotic fluid, meconium, and environmental contaminants, which may reflect the litter’s PRRSV status upon laboratory testing.
Objectives of the study described herein had a goal to further knowledge on the use of TF from stillborns in commercial sow herds for PRRSV detection. Objectives included evaluating 1) the incidence of stillborn piglets on the probability of PRRSV RNA detection within the litter, 2) the stillborn TF PCR-results as an indicator of PRRSV in liveborn piglets, and 3) the litter size as a risk factor for PRRSV
To investigate these objectives, samples were collected from two PRRSV-positive breeding herds (Herd A: 2,500-sow farm with a three-week batch farrowing system; Herd B: 4,500-sow farm with a weekly batch farrowing system). A total of 130 litters were sampled within 12 hours after farrowing: 66 litters without stillborn piglets and 64 litters with stillborn piglets. This totaled 1,723 liveborn and 105 stillborn piglets sampled. Regarding the litters’ parity, in Herd A, 14 were parity 1 sows, seven were parity 2, four were parity 3, and one was parity 4. For Herd B, all 104 litters came from parity 1 sows.
TF and intracardiac blood samples were individually collected from stillborn piglets. Tail blood swabs were individually collected from all liveborn littermates. Samples were individually tested for PRRSV RNA detection by RT-qPCR at the Iowa State University Veterinary Diagnostic Laboratory. Litters with ≤ 11 liveborn piglets were defined as small litters, whereas litters with ≥ 12 were defined as large.
Across all sample types, the percentage of PRRSV-positive litters was 22.3% (29/130 litters), of which 15.4% (20/130 litters) were tail blood swab PCR positive, 10% (13/130 litters) were intracardiac blood PCR positive, and 16.9% (22/130 litters) were TF PCR positive. At an individual animal-level across both herds, 3.4% (59/1,723 liveborn piglets) were tail blood swab PCR positive, 16.2% (17/105 stillborn piglets) were intracardiac blood PCR positive, and 26.7% (28/105 stillborn piglets) were TF PCR positive. The mean positivity of liveborn piglets within the litter was 5% but varied between 0 – 85.7%, while the total born (stillborn and liveborn piglets combined) was 4.6% (0 – 76.4%).
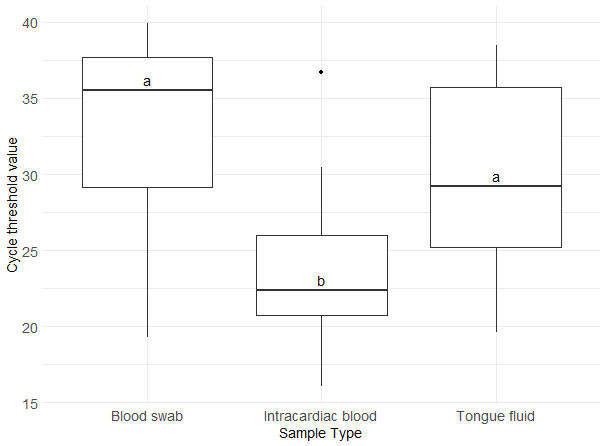
Regarding sample type Ct values, tail blood swabs had a median of 35.5 (interquartile range, IQR: 29.2-37.7), intracardiac blood a median of 22.4 (IQR: 20.7-26.0), and TF a median of 29.2 (IQR: 25.2-35.7) (Figure 1). Intracardiac blood exhibited the lowest median Ct value, which was significantly different from the median Ct values of the other sample types (P < 0.05).
In the model analysis, litter size and the presence of stillborn piglets were used as predictor variables, while litter PRRSV status was the response variable. Small litters had 12.2 times higher odds (P < 0.001) of having a PRRSV-positive piglet compared to large litters. Holding stillborn presence constant, there was a 51.9% probability of detecting PRRSV-positive piglets in small litters compared to 8.1% in large litters. Similarly, litters with at least one stillborn piglet had 12.5 times higher odds (P < 0.001) of having a PRRSV-positive piglet compared to litters without stillborn piglets. Holding litter size constant, there was a 52.2% probability of detecting a PRRSV-positive piglet when at least one stillborn was present, compared to 8.0% when no stillborns were present. No significant interaction between litter size and stillborn presence was found (P = 0.076).
When considering at least one TF PRRSV-positive result and litter size as predictors to assess the odds of having a PRRSV-positive liveborn littermate, litters with at least one TF-positive result had 17.6 times higher odds (P < 0.001) of having a PRRSV-positive liveborn piglet compared to litters without stillborn piglets. Holding litter size constant, there was a 63.6% probability of detecting PRRSV-positive liveborn piglets when at least one TF result is PRRSV-positive compared to 9.0% when stillborn were absent or 4.0% when TF result was PRRSV-negative. Similarly, small litters had 7.0 times higher odds (P = 0.002) of having a PRRSV-positive liveborn piglet compared to large litters. Holding the TF results constant, there was a 20.8% probability of detecting a PRRSV-positive liveborn littermate in small litters compared to 3.6% in large litters.
In conclusion, litters with stillborn piglets and small litter size have a higher probability of detecting PRRSV in the farrowing room. Moreover, TF from stillborn piglets are a reliable indicator of the PRRSV status of their liveborn littermates, indicating that TF can be an effective risk-based approach for PRRSV detection.
Veterinarians and producers are encouraged to collect TF, targeting stillborn piglets and small litters to increase the likelihood of detecting PRRSV. This risk-based sampling strategy within the first few hours post-farrowing enhances the effectiveness of PRRSV monitoring programs in breeding herds and supports timely interventions, such as McREBEL and cross-fostering. Lastly, while TF were individually collected in this study to test the hypothesis, collecting them as an aggregated sample, similar to PF, is recommended. This approach increases the likelihood of detecting the virus, allowing for a larger number of animals to be screened.

A study funded by the Swine Health Information Center Wean-to-Harvest Biosecurity Research Program, in partnership with the Foundation for Food & Agriculture Research and Pork Checkoff, sought to determine if an environmental enrichment (EE) device could be used to self-vaccinate pigs through natural behavior and reduce labor requirements. Led by Dr. John McGlone at Texas Tech University collaborating with Dr. Rebecca Robbins and Dr. Jessica Seate and a team of students, the EE device was developed to allow pigs to self-administer liquids, such as vaccines, through pig rooting, investigating, and natural play behaviors. Through measuring antibody response after self-vaccination compared to hand-vaccination, researchers investigated efficacy of vaccine delivery to pigs for erysipelas, iIleitis, influenza, and Mycoplasma hyopneumoniae. Researchers determined efficacy varies by pathogen, with erysipelas and ileitis vaccines showing similar efficacy between self and hand-vaccination.
Find the industry summary for project #23-052 on this page.
This study sought to determine if an EE self-administration device could deliver vaccines and generate robust antibodies in growing pigs against Lawsonia intracellularis (ileitis), Mycoplasma hyopneumoniae (Mhp), influenza A virus (IAV), and Erysipelothrix rhusiopathiae (erysipelas). For each vaccine trial, 36 growing pigs at approximately 200 pounds were utilized over a period of up to 49 days after vaccination. Oral fluids and serum antibodies were collected at baseline and post-vaccination to compare vaccine efficacy between the EE self-vaccination technology and traditional vaccine administration by a person. Assays for serum IgG and IgA were compared across three treatments, including 1) non-vaccinated controls, 2) pigs with individual vaccination by oral gavage or intramuscular injection, and 3) self-vaccinated group.
Pigs were raised in an experimental finishing barn similar to commercial conditions. The herd was naïve for PRRSV, PEDV, PEDCoV, TGEV, and Mhp at the time of the study. No Mhp, IAV or ileitis vaccines were being used in the sow herd; however, some pigs had background titers to ileitis prior to the study. All pigs received erysipelas bacterin at 21 days of age. Three pens (four pigs/pen) were utilized per treatment in a single barn. Each treatment group was accommodated in blocks of three adjacent pens, with contact possible through fencing within the treatment group. An empty pen was placed between treatment blocks to prevent contact between treatment groups. Pen configuration was to ensure no inadvertent exposure occurred between controls and vaccinates or between vaccination routes. The pig was the experimental unit in this study.
Pigs were randomly assigned to one of three treatments groups: 1) Control pigs that received no vaccine or exposure to the EE device, 2) Hand-vaccinated pigs receiving either IM injections of commercially licensed vaccines (Mhp and IAV) or 2.5 mL of vaccine delivered orally by hand with a syringe (erysipelas and ileitis), and 3) Self-vaccinated pigs exposed to a EE device attached to each pen which pigs could operate by pressing a panel with their snouts.
At the time of vaccine administration, 15 mL of MP (maternal pheromone) was sprayed on the front panel of the EE device to encourage pig interaction. When the panel was pressed, the device delivered a spray volume of up to 4 mL. Cameras were installed overhead, and network video recorders were used to record the self-vaccinated group to determine if all pigs received the vaccine. Sample collection was conducted at specific intervals post-vaccination to monitor antibody development.
Results for Mhp and IAV showed that self-vaccination with either antigen did not generate serum or oral fluid antibodies equivalent to the hand-vaccinated pigs. When erysipelas vaccines were tested, self-administered pigs developed both oral and serum antibodies equal to those of hand-vaccination. Pigs that self-administered the ileitis vaccine developed only oral fluids antibodies. It was determined that some vaccines have similar efficacy between self-administration compared to labor-intense individual vaccinations. However, other vaccines did not readily induce antibody synthesis.
Overall, findings suggest that self-administration of killed vaccines, such as IAV and Mhp, may not be effective when delivered via the EE device. Inactivated vaccines typically rely on systemic immunity and often require adjuvants or higher doses to enhance efficacy. These characteristics pose challenges for self-vaccination formats without precise dose controls. In contrast, live attenuated vaccines stimulate both systemic and mucosal immunity and can replicate locally at the administration site, making them more suitable for self-vaccination. Vaccine formulations or administration methods could be adapted to allow self-vaccination of select vaccines. Alternatively, an EE device may be modified to allow different methods of administration such as subcutaneous or intramuscular routes.
Using EE for self-vaccination of select vaccines has the ability to reduce labor requirements, eliminate the need for needles, provide benefits to animal welfare during immunization, and allow pen-level vaccinations or delivery of other animal health products. Self-vaccination via an EE device not only reduces labor demands but also offers a less stressful and more enriching experience for pigs compared to traditional hand-vaccination methods. This approach has the potential to improve vaccine compliance, biosecurity, and overall animal welfare in commercial swine operations using emerging technology being examined for application in the industry.

A study funded by the Swine Health Information Center Wean-to-Harvest Biosecurity Research Program, in partnership with the Foundation for Food & Agriculture Research and Pork Checkoff, recently defined biosecurity practices used across wean-to-harvest sites through an industry-wide questionnaire and developed a rapid-risk assessment tool for producers. Led by Dr. Gustavo Silva at Iowa State University, the study assessed current bioexclusion practices among a diverse group of producers across swine-producing states in the US. Further, the study developed methods that veterinarians, production managers, and producers could implement to improve on-farm biosecurity. Findings showed that biosecurity on wean-to-harvest sites is inconsistent across the industry and that tools for increasing biosecurity could include relatively simple practices such as bench entry.
Find the industry summary for project #23-029 here.
Two primary objectives of the study were to 1) assess the current bioexclusion practices used at wean-to-harvest sites across the US, ensuring a diverse group of producers from different swine-producing states are included and 2) develop a tool that veterinarians, production managers, and producers can use to assess biosecurity on their sites quickly.
Data to characterize current biosecurity practices was collected through a questionnaire completed by 21 herd veterinarians, representing production systems and independent producers. The questionnaire was developed with input from industry experts and was comprised of 69 questions on bioexclusion practices, covering site characteristics, vehicle movements, people movement, manure removal, water entry, and sanitation. A weighted method ensured the results reflected all respondents’ answers.
Results of the questionnaire included data representing 15.7 million pigs across 3,680 sites in 13 states. Of the 3,680 sites, 10.3% were nurseries, 52.9% were finishing sites, and 36.8% were wean-to-harvest sites. Most farms (93.3%) reported using all-in-all-out and mortality disposal was mostly off-site (65.3%). Close to half (47.3%) of all employees visited more than one site daily. While most sites have shower facilities (63.8%), fewer sites require employees to shower in or out (57.6% and 56.9%, respectively). Manure is removed about 1.5 times per year, often by third-party companies. Most sites rely on well water (87.7%) with most not performing any water treatment (64.7%).
Questionnaire results on transport biosecurity revealed trucks hauling pigs are generally washed and disinfected, with 100% of trucks hauling weaned pigs cleaned between loads. For feeder trucks, 60.9% are washed and 63.9% are disinfected between every load. For market hog trucks, 78.3% are washed, and 52% are disinfected between every load.
For developing the biosecurity assessment tool in Objective 2, researchers enrolled 139 wean-to-harvest sites to assess biosecurity practices and their relationship with lateral introduction of PRRSV, PEDV, PDCoV, and TGEV. Farms were required to be stable or negative for PRRSV and key enteric viruses, including PEDV, PDCoV, and TGEV, to participate. Participating producers were asked to complete a biosecurity questionnaire with 115 questions covering risk events, biosecurity management practices, herd demographics, truck sanitation, and farm location.
The 139 sites were across nine companies in six states, including 44 nurseries from three companies, 44 finishers from three companies, and 51 grow-finish sites. Findings demonstrated that PRRSV outbreak rates were highest in grow-finish sites (27/44; 61.4%), followed by wean-to-finish (27/51; 52.9%) and nurseries (15/44; 34.1%). No outbreaks of PEDV or coronaviruses were reported in nurseries or wean-to-finish sites, but grow-finish sites had a 2.3% break rate for coronaviruses and a higher rate for PEDV (11.4%).
Additional key findings included that nursery sites had 92% lower odds of reporting a PRRSV outbreak than finishers, and biosecurity practices like bench entry, truck washing, and downtime between loads reduced outbreak risk. Hauling animals with unknown status for PRRSV increased the odds of reporting an outbreak by 12 times, stressing the need for careful animal health monitoring before transportation.
While most sites involved in the study reported implementing biosecurity measures such as vehicle washing and employee training, researchers found gaps remain, especially in communication and compliance auditing. Information gained from Objective 2 revealed that nursery sites have a significantly lower risk of PRRSV outbreaks than grow-finish sites. This emphasizes the need for stronger biosecurity in the finisher phase. Simple, cost-effective measures like bench entry—where employees change footwear or clothing before entering different areas—can help reduce the spread of PRRSV and are relatively easy to implement. Researchers note more data is needed to refine biosecurity recommendations and help producers improve their practices, enhance surveillance, and build a more resilient industry.
Information collected through Objective 1 allowed researchers to understand and characterize the implementation frequency of bioexclusion practices on wean-to-harvest sites. Results are aligned with the reality of biosecurity measures being better established and more frequently implemented on sow farms than in wean-to-harvest populations. At wean-to-harvest sites, practices remain inconsistent and less rigorously enforced, where gaps in practices like hand hygiene, disinfection, and supply decontamination persist. As a result, the risk of pathogen introduction is heightened, revealing the need for stronger adherence to biosecurity protocols across this stage of production.
These findings highlight the need for targeted biosecurity measures, especially in the finisher phase, where the risk of outbreaks is higher. Results underscore the importance of implementing effective biosecurity practices, such as regular washing, downtime, and preventing sick animal transport, to mitigate the risk of PRRSV transmission.
SHIC actively monitors global swine diseases as part of its mission to enhance swine health through the identification of emerging disease threats. On March 7, 2025, Hungary reported its first foot-and-mouth disease outbreak in over 52 years at a dairy cattle farm in Kisbajcs, a town located near the Slovakian border. The outbreak affected a herd of 1,400 cattle, with classic FMD symptoms observed, including fever, excessive salivation, and blisters on the mouth, tongue, and hooves. On March 26, 2025, a second FMDV outbreak was reported in the same region of Hungary affecting a 3,000 head dairy herd. As part of the SHIC Global Swine Disease Monitoring Report, the team at the Center for Animal Health and Food Safety (CAHFS) has prepared this update of the FMDV situation in Hungary.
Key Dates in Hungary’s FMD Outbreak
March 3, 2025 – Clinical signs of FMD observed.
March 6, 2025 – Laboratory confirmation of FMD.
March 7, 2025 – Hungary officially reported the outbreak to the WOAH.
March 26, 2025 – Hungary confirms second FMDV outbreak.
Outbreak locations
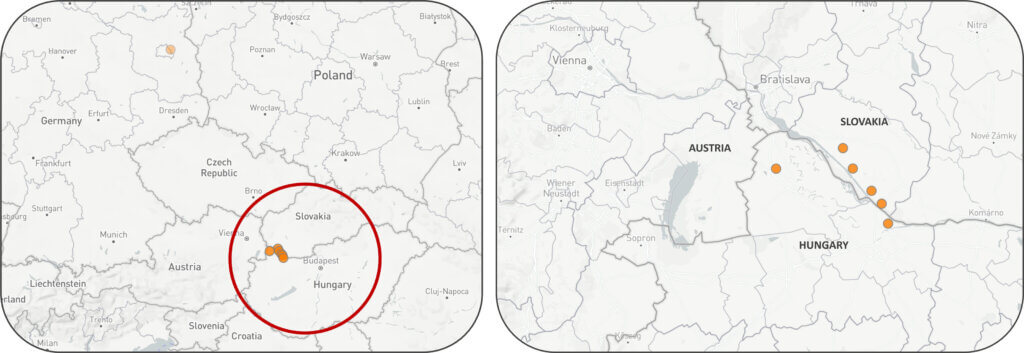
The initial outbreak occurred at a dairy farm in Kisbajcs, a town in northwestern Hungary located within 2 km of the Slovak border. The second outbreak occurred in Level, Gyor-Moson-Sopron county (less than 50 km (30 miles) from the first outbreak), the same region as the first outbreak near the Slovakian border.
Cause and Transmission: How and Why the Outbreak Occurred
The exact source of the virus remains unknown. Authorities are conducting epidemiological investigations to determine the source of the outbreak and assess the potential risk of further transmission. The Hungarian National Reference Laboratory has identified the FMD virus as serotype O. Genetic sequencing revealed that the strain shares 98–99% similarity with a virus isolated in Pakistan in 2017–2018, indicating a potential epidemiological link. Importantly, this strain is genetically distinct from the one detected in Germany in January 2025, confirming that the Hungarian and German outbreaks are not connected. Potential transmission routes include human clothing, shoes, exposure to infected animals, animal trade, and movement of animal products. Authorities are investigating the possibility of transmission to wildlife (e.g., wild boar and deer).
Measures Taken
The entire infected first herd (1,400 cattle) was culled. Hungary established a 3km protection zone and a 10km surveillance zone, extending into Slovakia. For the second outbreak, culling of infected cattle is anticipated to be completed soon, mandatory culling of pigs in the affected areas by March 31 due to their potential role in virus transmission, and grazing restrictions enforced along a 10 km strip near the border to prevent further spread.
Increased Surveillance
▪️ Susceptible herds within a 10km radius are being tested.
▪️ Blood samples are being collected from hunted wild animals in the region.
▪️ Zoos closed.
▪️ Hunting is prohibited within restriction zones to minimize the risk of virus spread.
▪️ Decontamination protocols say vehicles can only leave infected farms and burial sites after strict disinfection.
Movement Restrictions
▪️ National ban on the movement of susceptible cloven-hoofed species initiated.
▪️ Pigs, sheep, and goats can be transported for immediate slaughter outside affected counties.
▪️ Cattle remain under full movement restrictions.
International Trade Impact
▪️ 15 non-EU countries have temporarily banned imports of Hungarian meat products.
▪️ The EU has issued certification for continued trade within its member states.
Increased Surveillance
▪️ Susceptible herds within a 10km radius are being tested.
▪️ Blood samples are being collected from hunted wild animals in the region.
▪️ Zoos closed.
▪️ Hunting is prohibited within restriction zones to minimize the risk of virus spread.
▪️ Decontamination protocols say vehicles can only leave infected farms and burial sites after strict disinfection.
Overview of International Responses to Hungary’s FMD Outbreak
Romania
▪️ Enhanced surveillance of cattle, pigs, sheep, and goats.
▪️ Instituted mandatory reporting of illness or mortality in livestock.
▪️ Printed advisories posted in public places (e.g., city halls, veterinary clinics, churches).
▪️ Implemented stricter monitoring of animal transport from affected areas.
United Kingdom
▪️Banned imports of cattle, pigs, meat, dairy, and animal by-products from Hungary and Slovakia.
▪️ Restricted travelers from bringing meat, dairy, and animal by-products into the UK.
▪️ Conducting ongoing risk assessment with potential for further restrictions.
Kosovo
▪️ Put full import ban on live animals and animal products from Hungary in place.
▪️ Declared FMD-free status and enhanced disease monitoring.
Poland
▪️ Closed borders to imports of livestock, meat, dairy, and animal by-products from Hungary and parts of Slovakia.
▪️ Began sanitary border inspections at crossings with Slovakia and the Czech Republic.
▪️ Required mandatory disinfection of animal transport vehicles.
▪️ Deployed special vehicle disinfection gates at key border crossings.
Czech Republic
▪️ Banned transport of susceptible animals from Hungary and Slovakia (except for direct slaughter).
▪️ Conducting border inspections by veterinary authorities, police, and customs officials.
▪️ Requiring strict disinfection protocols for animal transport vehicles.
▪️ Restricted farm access for individuals recently in Hungary.
Ukraine
▪️ Banned imports of live animals, genetic material, and animal products from Hungary.
▪️ Monitoring the epizootic situation continuously.
Canada
▪️ Implemented import restrictions on live animals, raw meat, milk, and animal by-products from Hungary and Slovakia.
▪️ Issued border alerts and enhanced screening by Canada Border Services Agency.
▪️ Tracking shipments of pork and dairy products imported after February 2, 2025.
Australia
▪️ Suspended imports of meat and dairy products from Hungary and Slovakia.
Ongoing Concerns and Next Steps
The economic impact of trade restrictions is expected to last for several months. Authorities are monitoring wildlife and other livestock premises to detect potential further spread. Hungary will only be declared FMD-free after an extended period of testing and a confirmed absence of new cases.
With the recent addition of FMDV detection in Slovakia, the detection of FMD in three EU countries within a short timeframe raises concerns about biosecurity breaches, trade risks, and virus translocation within the region. This highly contagious disease requires continued vigilance and strict control measures to prevent further spread in Hungary, Slovakia, and other EU countries.
References
DEFRA Preliminary Outbreak Assessment – Foot and Mouth Disease in Hungary
Foot and mouth disease in Hungary: animals killed, bans in place, zoo closed
Government update about the spread of deadly disease in Hungary
Hungary reports first foot-and-mouth cases in 50 years
China has lifted some restrictions on German dairy product imports, German ministry says
Germany regains status as free of foot-and-mouth disease, ministry says
Hungary detects first case of foot-and-mouth disease in 50 years
Veterinary watchdog orders measures to prevent spread of FMD virus from Hungary to Romania
UK bans imports of some animal products from Hungary, Slovakia after foot-and-mouth case
Kosovo bans import of animals and their products from Hungary
Health checks at the border with Slovakia after detection of foot-and-mouth disease in Hungary
Czechia Bans Animal Transport from Hungary, Imposes Border Control with Slovakia
Foot-and-mouth disease: Ukraine has restricted imports from Hungary due to an outbreak of a dangerous animal disease
CFIA enforces new restrictions due to Foot and Mouth Disease in Hungry and Slovakia
Australia bans imports from two EU nations after foot-and-mouth detected
As part of its mission to identify emerging disease threats to the US pork industry, SHIC is monitoring the recent FMDV incursions in the EU, with Slovakia being the most recent country to report detection. Slovakia confirmed its first FMD outbreak in over 52 years at four cattle farms in the southern regions of Dunajská Streda and Komárno, near the Hungarian border. On March 30, 2025, a fifth cattle farm in the western Bratislava region was confirmed with FMD. Over 6,000 cattle are affected, with animals showing classic FMD symptoms such as fever, excessive salivation, and mouth and hoof blisters. Within the framework of the SHIC Global Swine Disease Monitoring Report, the team at the Center for Animal Health and Food Safety (CAHFS) has prepared this update of the FMDV situation in Slovakia.
The Slovakian outbreak follows recent cases in Hungary and Germany, making it the third confirmed FMD outbreak in Europe in 2025. Emergency measures, including movement bans, culling, and zoo closures, have been implemented while investigations continue to determine the source and contain further spread.
Timeline of Events: Key Dates in Slovakia’s FMD Outbreak
● March 21, 2025: The first three FMD outbreaks were confirmed at three cattle farms in Medveďov, Ňárad, and Baka (Dunajská Streda District), involving a total of over 2,760 susceptible animals (Baka: 1,301; Ňárad: 790; Medveďov: 670). A state of emergency was declared in the district.
● March 22–23, 2025: Vaccination completed at affected farms in Medveďov and Ňárad to reduce virus shedding before culling. Culling operations begin at the Baka farm, progressing at 300 animals per day.
● March 24, 2025: Ongoing culling operations and transport of slaughtered animals to disposal facilities.
● March 25, 2025: The fourth FMD outbreak was confirmed at a farm housing 270 cattle near Malá Lúč (Lúča na Ostrove), approximately 10km from Baka.
● March 30, 2025: The fifth FMD outbreak was confirmed at a large cattle farm housing 3,750 cattle in the western village of Plavecký Štvrtok, about 30 km northwest of Bratislava and not far from the Austrian and Czech borders. Authorities confirmed that this farm has a connection with one of the outbreaks in Hungary (Lével). Vaccination has started.
Outbreak Locations
The outbreaks confirmed in the southern part of the country are near the border with Hungary. These areas, particularly the Dunaszerdahely region, are major hubs for Slovakian livestock production, housing approximately 13,000 cattle and 128,000 pigs, and are now under intensive control measures due to their heightened vulnerability to disease spread.
1. Medveďov (Dunajská Streda District, Trnava Region)
● One of the first outbreak sites.
● Home to a large herd of milking cows.
● Managed by Naše Farmy, part of the J&T investment group.
2. Ňárad (Dunajská Streda District, Trnava Region)
● Also part of the Naše Farmy network.
● Symptoms were first noticed in mid-March.
3. Baka (Dunajská Streda District, Trnava Region)
● Operated by EXATA Group, owned by major Slovak business figures.
● The farm housed over 1,300 dairy cows and calves.
● A major milk producer, supplying 12 million liters annually prior to the outbreak.
4. Lúča na Ostrove (Dunajská Streda District, Trnava Region)
● Fourth confirmed outbreak, announced on March 25, 2025.
● Infected herd includes approximately 270 cattle.
● Located near previously affected areas, contributing to growing concerns about regional spread.
5. Plavecký Štvrtok (Malacky district, Bratislava Region)
● Fifth confirmed outbreak, announced on March 30, 2025.
● Affected site housing around 3,000 cows, 150 heifers, and 600 calves.
● Located approximately 30 km from Bratislava near the Czech and Austrian borders.
Cause and Transmission: How and Why the Outbreak Occurred
The exact source of the outbreak remains under investigation. However, authorities suspect the virus likely entered from neighboring Hungary, where an outbreak was reported in early March 2025, near the Slovak border. Affected farms are located in regions with dense livestock populations, making the area particularly vulnerable to fast and extensive transmission. The proximity of outbreak sites to Hungary and the interconnected nature of livestock trade and transport further contributed to the virus’s spread.
Measures Taken
In response, Slovakia has implemented strict emergency measures to contain the outbreak and prevent further spread:
● State of Emergency declared in the Dunajská Streda district.
● Nationwide ban on the movement of cloven-hoofed animals (cattle, pigs, sheep, goats, and farmed game), except for necessary transport within the same holding.
● International transport restrictions for susceptible animals through Komárno and Dunajská Streda districts.
● Culling of over 3,000 cattle, with operations ongoing and averaging 300 animals per day.
● 10,000 vaccine doses procured by the Ministry of Defence to support the emergency response and enhance outbreak control capacity.
● Vaccination at selected outbreak sites (e.g., Medveďov, Ňárad) to reduce virus shedding prior to humane culling.
● Disposal protocols in place:
○ Carcasses of visibly infected animals are incinerated at rendering plants.
○ Uninfected carcasses are buried at designated sites.
○ All transport containers are disinfected and sealed, with inspections at departure and arrival points.
● Over 1,500 police officers deployed to enforce movement restrictions and control zones.
● Fire and rescue services engaged in animal carcass transport, disinfection, and culling support.
● Installation of disinfectant fords and vehicle checkpoints on roads near infected premises.
● Closure of all zoos, circuses, and animal display facilities across Slovakia.
● Public forest access is banned in affected districts to reduce the risk of wildlife-mediated spread.
● Authorization for hunters to cull wild susceptible animals showing signs of disease, even outside regular hunting seasons.
● Coordination with the EU and WOAH, and an official request for financial compensation submitted to the European Commission.
EU experts acknowledged Slovakia’s response as best practice, strengthening the country’s eligibility for support.
Overview of International Responses to Slovakia’s FMD Outbreak
Poland
● Comprehensive import ban on live cloven-hoofed animals, meat, dairy, and by-products from Slovakia and Hungary.
● Enhanced inspections and disinfection at border crossings with Slovakia.
● Installation of disinfection gates at key border points (e.g., Barwinek).
● Trace-back investigation of over 400 animal shipments from Slovakia in the past month.
Czech Republic
● Initial ban on the transport of susceptible animals from Hungary and Slovakia (except for direct transport to slaughterhouses) imposed on March 7, lifted on March 18, then reinstated on March 21 following FMD confirmation in Slovakia.
● Import ban on livestock and animal products from affected areas of Slovakia (reinstated March 21).
● Border inspections at four major crossings, conducted by veterinary inspectors, police, and customs officers.
● Mandatory disinfection of transport vehicles and required documentation for biosecurity compliance (e.g., proof of vehicle disinfection, animal health certificates).
● Access restrictions to farms for individuals who had recently traveled to outbreak areas (e.g., Hungary, Slovakia).
● Deployment of 16 Czech firefighters and a mobile livestock culling unit to Slovakia to assist with containment, disinfection, and humane slaughter operations.
● Appeals to Czech farmers and agricultural unions to enforce strict biosecurity, including hygiene, disinfection, and bans on unauthorized entry into livestock facilities.
United Kingdom
● Banned imports of cattle, pigs, meat, dairy, and animal by-products from Slovakia and Hungary.
● Since 8 March, travellers to Great Britain are prohibited from bringing meat, meat products, milk, dairy products, certain composite products, animal by-products of pigs and ruminants, as well as hay and straw from Hungary and Slovakia.
Canada
● Immediate import restrictions on live animals, raw meat, milk, and animal products from Slovakia and Hungary.
● Border alert issued, and import certificates reviewed for compliance.
● Shipments arriving after February 2, 2025, are being traced and assessed.
Ukraine
● Ban on imports of live animals, genetic materials, and animal products from Slovakia and Hungary.
● Ongoing surveillance of the domestic epizootic situation to maintain disease-free status.
Australia
● Suspended imports of meat and dairy products from both Slovakia and Hungary due to FMD detection.
Ongoing Concerns and Next Steps
Slovakia’s FMD outbreak has raised serious concerns for both animal health and economic stability. With over 3,000 cattle affected and outbreaks confirmed near Hungary’s border, there is a high risk of regional spread, especially in areas with dense livestock populations. Authorities are closely monitoring wildlife and nearby farms to prevent further transmission. Despite containment efforts, the economic impact is growing, with estimated losses exceeding €10 million. Farmers are concerned about long-term consequences, including trade disruptions and livestock restocking challenges. Slovakia is coordinating with the European Commission to secure financial compensation and technical support. The government is also working with the WOAH to maintain transparency and ensure proper control measures.
Next steps include:
● Completing culling and disinfection operations.
● Expanding surveillance and testing in at-risk zones.
● Continuing cross-border coordination with neighboring countries.
● Finalizing damage assessments to access EU compensation mechanisms.
The situation remains dynamic, with authorities stressing the need for continued vigilance and swift regional cooperation to prevent further outbreaks. The distribution of FMD outbreaks is presented on Figure 1.
References
Emergency Measures for Foot-and-mouth Disease Enter into force
FMD confirmed in Slovakia, following outbreak in neighbouring Hungary
Slovakia reports first foot-and-mouth cases in 50 years
News digest: Slovakia confirms foot-and-mouth disease, begins culling livestock
Culling of Cattle at Baka Farm Continues at Rate of 300 Animals per Day
Foot-and-Mouth Disease Outbreak Prompts State Of Emergency In Slovakia
Cattle Vaccination in Medvedov and Narad Farms Completed, Culling Not Yet Begun
Slovakia to Request Compensation from EC for Foot-and-Mouth Disease
FMD: Slovakia and Hungary both see further outbreaks this week
Czech Republic Helping Slovakia Tackle Foot-and-mouth Disease
Czech firefighters join Slovakia’s fight against foot-and-mouth outbreak
Animals from Slovakia and Hungary Can Be Transported Again to Czech Republic
Czechia Bans Animal Transport from Hungary, Imposes Border Control with Slovakia
Czech Republic bans livestock imports from Slovakia after new foot-and-mouth outbreak
Czech Republic reinstates border controls with Slovakia following foot-and-mouth disease outbreak
Fifth outbreak in Slovakia

The Swine Health Information Center has released its 2025 Plan of Work Research Program Request for Proposals, allocating $1.5 million to fund targeted research addressing critical swine health priorities. Proposal submission deadline is April 30, 2025, at 5:00 pm CST. This initiative fulfills SHIC’s mission of mitigating emerging disease threats through strategic investments in research aligned with its five strategic priorities: improving swine health information, monitoring and mitigating risks to swine health, responding to emerging diseases, surveillance and discovery of emerging diseases, and swine disease matrices.
2025 SHIC Plan of Work Proposal Template & Instructions
With $1.5 million in funding available, individual awards are anticipated to range from $50,000 to $150,000. Proposals exceeding this range require strong justification. Proposals are expected to clearly address at least one of the identified priorities and demonstrate the project’s urgency, value to US pork producers, and efficient use of funds. Collaborative projects involving industry, academia, and public/private partnerships are highly encouraged. Projects should be completed within 12 months, with sufficient justification required for longer durations. Proposals will undergo a competitive review process by a Review Task Force, who will provide funding recommendations to the SHIC Board of Directors.

This month’s Domestic Swine Disease Monitoring Report brings information about the seasonal decrease in PRRSV case positivity, mainly in wean-to-finish sites. However, Iowa and North Carolina had higher PRRSV case positivity compared to historical data. Also, Iowa has a high diversity of strains circulating, with 26 PRRSV variants detected in 2025. PRRSV lineage 1C.5 detections continue to increase, reaching more than 600 detections in 2025. The report includes a bonus page about TGEV, including details on four years without a single positive case. More than 176,000 cases were submitted across six VDLs, with more than 500,000 RT-PCRs performed and not a single PCR-positive TGEV case. PEDV and PDCoV facilities’ case positivity decreased in March, but overall positivity was above the expected in Indiana. Mycoplasma hyopneumoniae case positivity in sow farms decreased, maintaining the promising trend of reduced activity of this pathogen in sow farms. PCV2 case positivity by PCR remains high in the wean-to-market category. However, the PCR Ct values are not as low as in previous periods of PCV2 activity. In the podcast, Dr. Lauren Glowzenski, Pipestone Veterinary Services, discussed the impact of PRRSV diversity, the hypotheses for TGEV disappearance, and strategies for handling pathogen co-infections in farms.

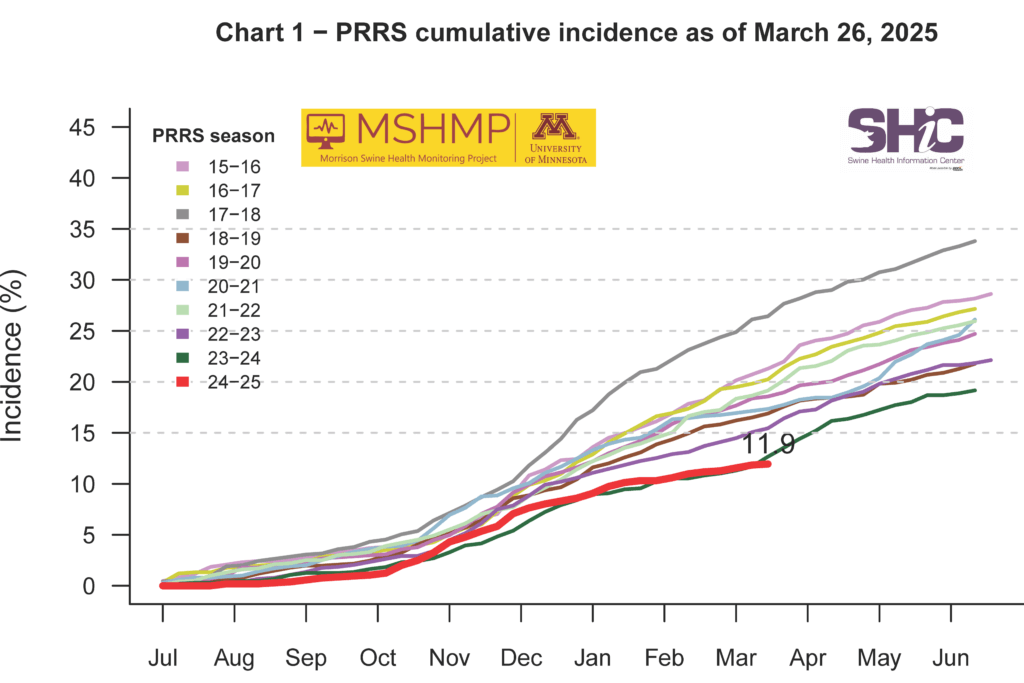
PRRS Cumulative Incidence for MSHMP Beginning July 1, 2009
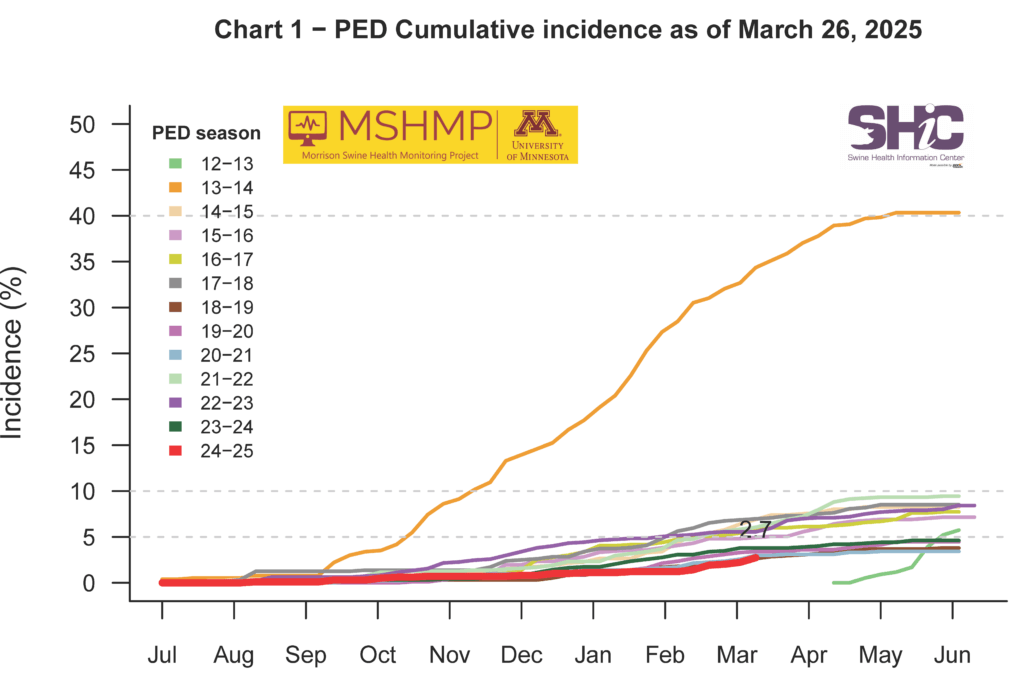
PEDV Cumulative Incidence for MSHMP Beginning May 1, 2013