
The Swine Health Information Center, launched in 2015 with Pork Checkoff funding, protects and enhances the health of the US swine herd by minimizing the impact of emerging disease threats through preparedness, coordinated communications, global disease monitoring, analysis of swine health data, and targeted research investments.

SHIC Executive Director Dr. Megan Niederwerder and Foundation for Food & Agriculture Research Scientific Program Director Dr. Jasmine Bruno discuss the tremendous response to the organizations’ joint call with Pork Checkoff for research proposals on H5N1 risk to swine.

Research provides critical information and drives innovation to help pork producers as they face emerging disease challenges in their swine herds. Now available, the Swine Health Information Center has released its 2025 Plan of Work Research Program Request for Proposals, allocating $1.5 million to fund targeted research addressing critical swine health priorities. This initiative fulfills SHIC’s mission of mitigating emerging disease threats through strategic investments in research aligned with its five strategic priorities: improving swine health information, monitoring and mitigating risks to swine health, responding to emerging diseases, surveillance and discovery of emerging diseases, and swine disease matrices.
The SHIC 2025 Research Program RFP, a strategic investment in swine health, fosters innovation and collaboration to protect the US swine herd. By addressing critical research priorities, SHIC provides tools to enhance preparedness, prevent disease introduction, and ensure the long-term sustainability of the swine industry. Researchers are encouraged to submit proposals that contribute to the advancement of swine health and the protection of this vital agricultural sector.
SHIC’s Strategic Priorities and the 2025 Plan of Work
The 2025 Plan of Work, developed with stakeholder feedback and approved by the SHIC Board of Directors, provides a framework for targeted research investments. The SHIC 2025 Research Program RFP invites qualified researchers to submit proposals that specifically address targeted research priorities outlined in the Plan of Work. With $1.5 million in funding available, individual awards are anticipated to range from $50,000 to $150,000. Proposals exceeding this range require strong justification.
Proposals are expected to clearly address at least one of the identified priorities and demonstrate the project’s urgency, value to US pork producers, and efficient use of funds. Collaborative projects involving industry, academia, and public/private partnerships are highly encouraged. Projects should be completed within 12 months, with sufficient justification required for longer durations. Proposals will undergo a competitive review process by a Review Task Force, who will provide funding recommendations to the SHIC Board of Directors.
Funding timely research is essential for SHIC to provide novel project outcomes that drive action for producers and veterinarians. This initiative aims to enhance US preparedness, prevent emerging disease entry, and enable rapid mitigation of pork production impacts. The SHIC 2025 Plan of Work Research Program RFP represents a significant commitment to funding research that directly addresses industry needs and strengthens the US swine herd’s resilience.

Glaesserella australis, a newly recognized Gram-negative bacterium species, was isolated from the lung lesions of pigs in Australia in 2018 and in two swine herds in Ontario, Canada in 2023. In response, the Swine Health Information Center funded a study led by Dr. Nubia Macedo, Iowa State University, to investigate the detection and characterization of this potential emerging swine pathogen in US clinical samples. Diagnostic tools developed through this project were used to screen historic isolates and current clinical samples. All US swine samples tested to date have been negative for this emerging bacterium. The tools developed herein will contribute to accurate detection of G. australis and help inform understanding of this potential pathogen.
Find the industry summary of project #23-053 here.
G. australis causes clinical signs and pathologic lesions that appear very similar to that caused by Actinobacillus pleuropneumoniae. Because no methods were available to detect G. australis in swine samples at US VDLs, ISU investigators hypothesized that infections may have been misdiagnosed as A. pleuropneumoniae. The overall goals of this study were to develop and optimize diagnostic tools to detect the presence and prevalence of G. australis in the US. Aligned with SHIC’s mission to protect the health of the US swine herd, surveillance and discovery of emerging diseases such as G. australis is critical.
In the affected Australian farms, most pigs had no signs of respiratory disease but there was increased pleurisy and pulmonary abscesses at the abattoir. On one farm, three affected pigs, aged 12, 16, and 20 weeks old, presented with cough and reduced growth rates followed by sudden death with cyanosis of extremities. Pulmonary lesions in these pigs affected up to 50% of lungs, characterized by multifocal necrotizing and fibrinosuppurative bronchopneumonia. In these cases, G. australis was isolated in pure culture. Other farms positive for G. australis had co-infections with other pathogens, including PCV2, A. pleuropneumoniae, Pasteurella multocida, Glaesserella parasuis, Streptococcus suis and Mycoplasma hyopneumoniae.
To date, there are no reports of detecting G. australis in US swine samples. However, G. australis was detected for the first time in two swine herds in Ontario, Canada, in 2023 when the bacterium was isolated from the pericardium and lung of one pig in each herd. Co-infections in the Canadian swine were present and included Streptococcus suis, Pasteurella multocida, Actinobacillus porcitonsillarum, IAV, and PRRSV.
In the US, the identification of a potential G. australis case could have been missed by VDLs for two reasons, including similarities to other pathogens, such as Glaesserella parasuis and Actinobacillus pleuropneumoniae, and a lack of diagnostic tools to accurately identify G. australis. Researchers sought to develop accurate diagnostic methods and increase awareness among producers, veterinarians, and diagnosticians on this potential pathogen threat.
Specifically, the objectives of the study were to 1) phenotypically characterize historical ISU VDL Actinobacillus sp. isolates using traditional biochemical methods and whole genome sequencing (WGS) and 2) develop and optimize diagnostic tools for G. australis detection within US swine herds including ISH and real-time PCR.
First, sequencing of historical isolates with similar characteristics was attempted at the ISU Veterinary Diagnostic Lab, but no US sample sequence was found to be a match for G. australis. Twenty-one isolates from the ISU VDL inventory were selected for further identification with WGS revealing that 19 isolates were closely related to Actinobacillus minor or A. porsitonsillarum. Final identification of the remaining two isolates in underway. For testing of future submissions, the G. australis reference strain was added to ISU VDL’s sequencing platform and to its MALDI-TOF database for real-time screening of clinical samples.
Second, a PCR test was developed, validated and is now available for further screening and diagnostic purposes. Using the TaqMan-MGB-based primer/probe sequences, the RT-PCR proved to be highly specific and sensitive. An ISH assay was developed and is currently undergoing validation to detect this organism in tissues associated with lesions. Once the ISH assay is available, it will allow the screening of G. australis directly in affected tissues, as well as detect concurrent pathogens such as G. parasuis, A. pleuropneumoniae, and P. multocida.
Since January 2024, ISU VDL staff have screened for G. australis through MALDI-TOF and all samples have been confirmed as negative for this new swine pathogen. Nevertheless, the tools described here will contribute to the G. australis screening process in the US and help improve understanding of its prevalence and pathogenesis. In addition to the ongoing MALDI-ToF screening, the RT-PCR is available for surveillance and a sequencing pipeline is in place to quickly and accurately confirm any suspect isolates identified.
The emergence of G. australis highlights the importance of ongoing disease surveillance and the development of accurate diagnostic tools. Producers should be aware of this potential new pathogen and consult with their veterinarians if they observe respiratory clinical signs in their pigs. The availability of these new diagnostic tests will improve the ability to detect and manage G. australis if present in the US swine population, protecting herd health and productivity.

The Swine Health Information Center, in collaboration with the American Association of Swine Veterinarians, hosted a webinar highlighting practical approaches for transportation biosecurity on February 18, 2025. Presenters provided applied information on transport biosecurity strategies for swine disease prevention and control. Topics included alternative livestock trailer cleaning methodologies to manage the risk of PEDV introduction, cost effective trailer cleaning and disinfection based on PEDV prevalence and system connectivity, rerouting vehicles as an alternative strategy for transport biosecurity, and implementation of routine market haul transport biosecurity for PEDV control on farm.
View the webinar here.
Presenters offering their expertise included Dr. Edison Magalhaes, Iowa State University, Dr. Ben Blair, University of Illinois Urbana-Champaign, Dr. Gustavo Machado, North Carolina State University, and Dr. Pete Thomas with Iowa Select Farms. Sharing of information and strategies gained from the SHIC-funded Wean-to-Harvest Biosecurity Research Program assists producers and their veterinarians in developing the best strategies and practices for biosecurity within and between their farms or systems.
First, Dr. Edison Magalhaes reviewed key results from the US SHIP Transport Biosecurity survey identifying the ongoing need to assess and validate different methods of trailer decontamination for PEDV control. Specifically, the trailers hauling growing pigs for run out loads were shown to be less frequently washed between loads and present an opportunity for improved sanitation.
Dr. Magalhaes went on to share experimental data from an evaluation of trailer cleaning methods for the ability to remove PEDV from a contaminated trailer. The project used a small-scale trailer model to evaluate various washing and decontamination methods. Trailers were uniformly contaminated with slurry containing PEDV-infected feces. Five cleaning methods were assessed including scrape and bake, high volume wash, power wash with disinfectant, and two control treatments. To mimic the transmission and spread of PEDV via personnel, foot traffic between the trailer and the farm site was simulated and samples were collected for PCR and bioassay testing pre- and post-cleaning. Results showed both high volume wash and power wash with disinfectant were most effective in reducing PEDV trailer contamination.
Dr. Blair shared information on determining the best practices that balance disease control and economic feasibility across different swine production scenarios. Two system types were evaluated using computer simulations to model swine production under various conditions. The first scenario focused on a single production system with 24,000 sows across eight sites. The second scenario evaluated a region with 24,000 sows divided into 4 geographically related systems but operationally independent. Simulations evaluated how washing different proportions of trailers at an 80% washing efficacy rate would affect the spread of PEDV.
Scenarios were conducted with PEDV prevalence levels ranging from low (5%), moderate (10%) and high (20%) to see how PEDV prevalence impacted the effectiveness of trailer washing on disease reduction. This study found washing 100% of trailers resulted in the lowest mean number of infected premises in the mixed flow system. In the geographically segregated system, washing zero trailers was the most cost-effective strategy. For interconnected systems or during high PEDV prevalence periods, thorough washing of all trailers is essential. However, in more isolated systems or when PEDV prevalence is low, producers could save costs without compromising biosecurity by washing fewer trailers. Overall, the study showed that optimal strategies depend on individual production settings and regional disease prevalence pressures within that system.
Dr. Machado shared information from recent work examining the role of vehicles in transmitting swine diseases and the potential to rerouting vehicles as an alternative disease mitigation strategy. To mitigate disease transmission events, vehicles undergo cleaning and disinfection (C&D) procedures but C&D effectiveness and the frequency of C&D between farm visits is often unknown. Consequently, relying solely on vehicle C&D may be insufficient to stop the spread of diseases, and supplementary strategies are needed to prevent disease transmission events by contaminated vehicles.
Using GPS data from farms to trace vehicle movements in several pig-dense areas and incorporating PRRSV and PEDV infection status of commercial swine farms, Dr. Machado’s study simulated vehicle movements for 1 week. Vehicle movements included visits to farms, slaughterhouses, feed mills, and parking areas. Rerouting vehicles based on risk decreased deliveries per vehicle and the connectivity among vehicle networks while increasing C&D visits and the distance traveled. Given the severe economic impact of PRRSV, PEDV, and other endemic infectious diseases on swine production, the costs and logistics of a vehicle rerouting system will require a close economic examination. The potential health benefits of reduced disease transmission should be evaluated across traditional versus rerouted vehicle movement schedules.
Dr. Pete Thomas shared the history and challenges associated with PEDV within Iowa Select Farms and the impact of implementing increased transport biosecurity. He noted the gilt acclimation program for PEDV ended in 2022, resulting in a subsequent increase in sow farm outbreaks. Prior to 2023/2024, market hog trailers were washed every fourth load using volume flush and disinfectant. The estimated cost of a PEDV outbreak using 2018 – 2022 data from their system was calculated at $1.15/pig annually due to losses in weight gain and nursery mortality rates.
In week 46 of 2023, the company started washing all market trucks and noted a significant reduction in PEDV outbreaks including farm rebreaks. The washing of all market trucks reduced the break rate from 6% to 2% in 2023-2024. After focused efforts were made for transport sanitation, a significant reduction in outbreaks was noted from 20% (2018-2019) to 2% (2023-2024). In summary, the study showed an 11x reduction in PEDV breaks in marketing after implementing consistent trailer washing. Farms also saw a reduction in rebreak rates in weaned and feeder pigs.
Transportation biosecurity and sanitation play a key role in reducing disease spread and improving the overall health and productivity of swine herds. While the information presented offers valuable guidance, producers are encouraged to adapt biosecurity strategies to their unique production contexts and continuously refine their practices based on ongoing assessments and expert advice. Decontamination strategies should be tailored to address the disease risk within individual farms and production systems.
Additional steps that can enhance biosecurity include performing an assessment of current biosecurity measures with a focus on areas of improvement, tailor decontamination strategies based on disease prevalence in the area considering efficacy and cost, evaluation of rerouting strategies for disease prevention, and then adjusting strategies as needed depending on emerging data and best practices.

Evaluating whether post-mortem specimens such as tongue tip fluids (TTF) can serve as a viable tool for disease surveillance during the post-weaning stages of pork production is needed. To help fill this knowledge gap, the Swine Health Information Center funded a study to determine if TTF can aid in detection of key pathogens affecting swine during the grow-finish phase, including PRRSV, porcine circovirus, porcine parvovirus, Lawsonia intracellularis, and influenza A virus. Led by Drs. Cesar Corzo and Marcello Melini at the University of Minnesota, the objectives of the study were to 1) assess the sensitivity and specificity of PRRSV detection in TTF compared to other specimens including intracardiac blood, oral/nasal swabs, rectal swabs, and superficial inguinal lymph nodes and 2) characterize the detection of PCV2, PCV3, PPV1, PPV2, Lawsonia intracellularis, and IAV across all sample types. Overall, most pathogens were detected in TTF indicating that this specimen can provide valuable post-mortem information during a diagnostic investigation.
Find the study summary of project #23-063 on this page.
In the US, most postweaning swine health monitoring sampling relies on oral fluid collection. As labor constraints are a concern, assessing whether easy-to-collect post-mortem samples can provide value for diagnosis in these production phases is needed. Furthermore, finding practical and time-efficient methodologies to monitor health during the post-weaning stages provides value to swine health and production. With these potential advantages, industry adoption of TTF collection can help improve understanding of disease occurrence and dynamics.
Understanding whether TTF can effectively screen pathogens commonly found in postweaning pigs provides valuable insights for producers, offering potential solutions to rapidly respond and improve herd health management when conventional sampling methods are unavailable or impractical. By comparing the diagnostic performance of alternative specimens to traditional serum sampling for PRRSV and other pathogens through PCR, this research seeks to offer producers practical, cost-effective options for post-weaning disease surveillance.
To complete this study, samples were collected from two growing pig farms in Minnesota. The first farm, a 2,400-head wean-to-finish farm undergoing a PRRSV outbreak, was visited when pigs were five and 11 weeks of age. The second farm was a 3,300-head finishing site undergoing a similar health challenge as farm 1; this site was visited when pigs were 15 weeks of age. During each farm visit, a total of 30 dead pigs were sampled, resulting in a total sample size of 90 pigs. From each pig, TTF, intracardiac blood (IC), oral/nasal swabs (ONS), rectal swabs (RS), and superficial inguinal lymph node (SILN) samples were collected. Samples were tested for specific pathogens against a gold standard sample and then compared with TTF results.
All TTF samples were tested individually by PCR for all pathogens listed above. All specimens were tested individually for PRRSV. Additionally, ONS were individually tested for IAV; RS were individually tested for PPV1, PPV2, and Lawsonia intracellularis; and SILN were individually tested for PRRSV, PCV2 and PCV3. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated for PRRSV TTF and IC serum as the gold standard. The proportion of PCR positive results from specimens tested for other pathogens was compared by descriptive statistics. Most pathogens were detected at least once in TTF with Ct values ranging from 11.6 to 39.8.
For PRRSV, the best results across all specimens were in samples collected at 11 weeks and are shown in the table.
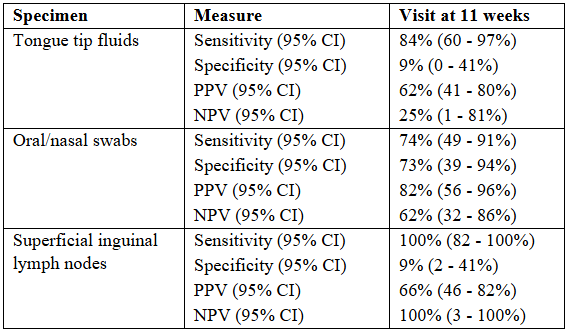
For other pathogens, PCV2 was detected in 43% of TTF and 11% of SILN samples whereas PCV3 was not detected in any sample. PPV1 was detected in 1% of TTF and 0% of RS whereas PPV2 was detected in 97% of TTF and 61% of the RS samples. Lawsonia intracellularis was detected in 6% of TTF and 0% of the RS samples. IAV was detected in 38% of TTF and 38% of the ONS samples.
Study data demonstrated that most pathogens were detected on TTF samples during the three different ages, indicating that this specimen can provide valuable post-mortem information during a diagnostic investigation. Detection of pathogens in TTF could be the result of shedding or environmental contamination which should be considered when practitioners are interpreting results. Further investigation is required to best report the diagnostic performance of the other alternative specimens utilized in this study. While a complete and exhaustive collection of multiple clinical specimens from different body systems remains the standard in diagnostic investigations, TTF provide an easy-to-collect sample type with low labor requirements and valuable diagnostic test results.
Swine viral pathogens such as PRRSV and PEDV pose significant economic challenges to the swine industry. The Swine Health Information Center funded a study to investigate novel diagnostic tools for differentiating infectious versus non-infectious swine viruses to use on-site for real-time detection. The study, led by Dr. Yi Lu at University of Texas Austin, aimed to develop a novel method for differentiating intact viruses versus those virus particles rendered noninfectious by disinfection. Novel diagnostic tools such as the one investigated in this study could provide valuable information to pork producers and veterinarians by evaluating cleaning and disinfection efficacy after a viral disease outbreak.
Find the industry summary of project #22-003 here.
While diagnostic methods such as PCR and cell culture are available to detect viral genetic material and assess infectivity, respectively, inherent limitations to these tests include required sample pretreatment and a skilled operator for processes performed on costly equipment. Further, the tests require hours to days to be conducted in a professional laboratory, making these tests unsuitable for on-site detection. The goal of the study described herein was to develop a novel method for direct detection of intact viruses without any sample pretreatment using a handheld portable meter.
The novel method under investigation is based on DNA aptamers that can be selected to bind and differentiate infectious swine viruses from noninfectious and other viruses. By immobilizing the aptamers into a nanopore, only infectious PRRSV or PEDV produce electrochemical signal changes and thus can be detected and quantified using a handheld meter. This innovative direct detection method utilizing DNA aptamers integrated into solid-state nanopores has previously demonstrated success in detecting a wide range of human viruses, including and human adenovirus and SARS-CoV-2.
There were two primary objectives of the study to apply the technology to swine viruses:
To conduct this study, PRRSV and PEDV samples were prepared in the laboratory of Dr. Ying Fang at the University of Illinois at Urbana-Champaign. A portion of the PRRSV and PEDV samples were rendered noninfectious by adding binary ethylenimine or UV light and then purified. Researchers conducted 10 SELEX (systematic evolution of ligands by exponential enrichment) experiments under varying conditions to optimize aptamer selection for PRRSV and PEDV. The SELEX process is designed to identify aptamers which are short, single-stranded DNA or RNA molecules that bind selectively to target viruses.
In previous work by this research group, SELEX projects targeted other viruses, such as human adenovirus and SARS-CoV-2. Using lessons learned from previous work, researchers optimized aptamer selection conditions by varying factors such as virus concentration, purity, and reaction times. Initial findings indicated that virus purity was a critical factor affecting the success of selections. Researchers focused on conducting SELEX experiments using ultra-centrifugally purified PRRSV and PEDV. This approach yielded promising results where shifts in melting temperatures that mirrored previous successes with other viral targets were observed.
After performing NGS on these more refined samples, data was analyzed, and several promising DNA sequences were obtained. Biochemical studies identified aptamers with the highest selectivity for either infectious PRRSV or PEDV. Currently, researchers have identified three different aptamer candidates for PRRSV. First, candidate 5 demonstrated binding to active PRRSV II and inactive PRRSV II, with no binding to active PEDV or active PRRSV I. Second, candidate 7 exhibited binding to active PRRSV II but no binding to inactive PRRSV II, active PEDV, or active PRRSV I. Third, candidate 9 showed binding to both active PRRSV II and active PRRSV I, while no binding was observed with inactive PRRSV II or active PEDV.
Despite these promising results, challenges with reproducibility and consistency in binding assays occurred in this initial pilot study. Additional optimization or complementary techniques may be required for field applications. The insights gained from this study underscore the importance of sample purity and the need to further enhance aptamer selectivity to improve detection reliability in applied settings. Further optimization of the DNA aptamer sequences, and assay conditions are required to develop this system into a practical sensor for on-site and real-time detection.
Through this pilot project, valuable lessons about the complexities of aptamer selection and the importance of optimizing experimental conditions have been gained for PRRSV and PEDV, two of the most important viruses affecting US swine. Moving forward, researchers aim to refine aptamer candidates to improve specificity and reliability for detecting viruses in porcine samples. These insights will guide future research efforts, contributing to the development of more effective diagnostic tools for the pork industry.

Porcine circoviruses, including porcine circovirus 2 and porcine circovirus 3, have been associated with clinical syndromes in swine, resulting in significant economic losses. To better understand the epidemiology and clinical relevance of PCV2 and PCV3, the Swine Health Information Center helped fund a study to analyze diagnostic data collected between 2002 – 2023 for PCV2 and PCV3 from six US veterinary diagnostic laboratories. The research, led by Drs. Giovani Trevisan and Daniel Linhares at Iowa State University, aimed to evaluate the macroepidemiological trends of aggregated PCV2 and PCV3 PCR data over time, establish the real-time capacity to rapidly identify changes in PCV2 and PCV3 detection patterns, and investigate the association between PCV-positive PCR cycle threshold (Ct) values and confirmed PCV disease diagnosis in tissues.
Guilherme Cezar, the graduate student working with the team, reported a decrease in the percentage of PCV2-positive submissions after introducing a commercial PCV2 vaccine in 2006 and a resurgence in positivity after 2018. The 2018 resurgence was primarily in breeding herds associated with an increased number of processing fluid sample submissions. PCV3 detection was more frequent in adult/sow farms, while PCV2 was more frequently detected in the wean-to-market category. An interpretative Ct cutoff of 22.4 for PCV2 was associated with a high probability of confirming a PCV2 disease diagnosis through histopathology. For PCV3, the interpretative Ct cutoff with the highest performance was 26.7.
See the published study with references here. Visit the SDRS site here.
Porcine circovirus-associated disease (PCVAD) can cost producers an average of $3–4 per pig in economic losses, demonstrating the importance of monitoring and controlling these pathogens in swine farms. Due to the complexity of monitoring PCV clinical cases, there is a need to develop tools to reveal and monitor changing patterns of PCV2 and PCV3 detection in swine farms. This study used aggregated PCR cases reported by VDLs for PCV2 and PCV3 detection to unravel the megatrends of these viruses in the US over the last two decades.
PCV2 and PCV3 submissions from 2002 to 2023 were collated using distinct accession IDs. PCR results reported by the VDLs (positive, negative, suspect, or inconclusive) were used to establish the final case result in the database. For cases to be considered positive, at least one sample within the case was required to be PCR-positive. Alternatively, negative cases had to have negative PCR testing results across all samples. Suspect and inconclusive cases were reported according to each laboratory criteria.
A variable age category was created based on the farm type, age unit, and age variables provided by VDL submissions. If the farm type was provided, the age category was assigned based on this variable (e.g., suckling piglets, breeding herd, nursery, grow-finish, replacement, boar stud). When the farm type was not provided, the age category was established based on the age of the animals (0–21 days as suckling piglets; 22–63 days as nursery; 64–200 days as grow-finish; >200 days as adults). Then, the age categories were aggregated into phases: adult/sow farm (breeding herds, replacement, boar stud, suckling piglets, and adults) and wean-to-market (nursery and grow-finish). Submissions lacking information regarding farm type, age, or age unit were categorized as unknown.
The Ct values of the PCR results were aggregated using the positive samples within a laboratory submission to calculate the average, minimum, and maximum PCR-positive Ct values. For example, a submission with three positive samples for PCV2 with Ct values of 34, 33, and 23 had an average, minimum, and maximum Ct value assigned as 30, 23, and 34, respectively. The number of positive samples was assigned based on distinct sample IDs within a case. In cases where the sample ID had more than one result, such as retesting the same sample, the most recent reported result date was retained.
The final PCR database comprised 154,984 PCV2 cases from 2002 to 2023. The first PCV3 PCR data was recovered in 2016 and had a total of 49,975 cases tested up to December 2023. The generated and aggregated information was made publicly available in an online visualization platform at the SDRS website.Data analysis of PCR test results, sample type, and age group revealed several key findings. PCV2 cases averaged nearly 6,000 annually from 2002 to 2017, rising to over 10,800 annually from 2018 to 2023. While the percentage of PCV2 positive cases peaked in 2006 (75%), it dropped significantly by 2011 (27%) and then rose again, averaging around 41% from 2018 to 2023. PCV3 testing became more frequent after 2018, with positive case percentages averaging around 50% per season. The study also noted an increasing trend of concurrent PCV2 and PCV3 detection since the introduction of multiplex PCR testing, which also contributed to the increase in the frequency of PCV3 cases reported after 2018.
Sample types submitted for testing also changed over time. While tissue samples were initially the most common for PCV2, processing fluids became the most frequently submitted sample type for both PCV2 and PCV3 after 2018. The age category unknown decreased significantly over time due to improved data capture.
Further, tissue diagnostic data from one VDL between 2019 and 2023 was used to correlate PCR Ct values with confirmed PCV2 and PCV3 disease diagnoses. The analysis, which considered only cases with tissue evaluations by diagnosticians, aimed to establish interpretative Ct cutoffs. Using a logistic regression model, a Ct cutoff of 22.4 for PCV2 and 26.7 for PCV3 was associated with a high probability of confirming a disease diagnosis through histopathology. These cutoffs represent the point where the likelihood of disease, despite a positive PCR result, decreases significantly. Specifically, Ct values above these cutoffs suggest that while the virus may be present, it is less likely to be contributing to clinical disease.
This study unraveled the macroepidemiological aspects of PCV2 and PCV3 in the US swine population. Additionally, a PCV2 monitoring tool based on the interpretative Ct cutoff results was implemented into the SDRS project and added to monthly PDF reports for continuous updates. Monitoring average Ct values of PCV2 tissue cases in the US helps to alert the industry when the average Ct of submissions is below 22.4 and can aid producers and veterinarians in recognizing higher PCV2 activity in the field. The study also sheds light on PCV3 detection trends, contributing to further investigations regarding virus dynamics. Detection megatrends revealed under this work can be used to further guide research questions and design experimental or field-based trials to explore and clarify root causes of secular trends in PCV2 or PCV3 detection.
This information will help pork producers and their herd veterinarians understand these pathogens and aid in disease management decision making. This research provides valuable insights into the evolving epidemiology of PCV2 and PCV3 in US swine herds. The established Ct cutoffs offer a practical tool for interpreting PCR results and may aid in disease management decisions. The study also highlights the importance of ongoing surveillance and data analysis to understand the complex dynamics of these economically significant swine pathogens.

This month’s Domestic Swine Disease Monitoring Report incorporates a new category for PEDV and PDCoV PCR detection: facilities. This new category includes PCR tests from truck washes, vehicles, and packing plants. With this new facility category, SDRS can monitor enteric coronavirus activity from outside the farms. The report includes a bonus page explaining the latest charts for the facility category. PEDV and PDCoV case positivity increased in the new facility category. Also, PEDV and PDCoV overall positivity was above expected in Indiana. The report brings information about the decreased case positivity of PRRSV in wean-to-finish sites. However, PRRSV lineage 1C.5 is spreading rapidly in 2025, exhibiting a historical record compared with the beginning of previous years. Influenza A case positivity increased in all age categories, with the highest positivity coming from wean-to-finish sites (33%). In the podcast, Dr. Tyler Bauman, herd veterinarian with The Maschhoffs, discussed health management strategies in finishing sites for control and elimination of Mycoplasma hyopneumoniae, IAV, PEDV, and PRRSV, including the different clinical implications of PRRSV L1C.2 and L1C.5

The March Global Swine Disease Monitoring Report includes news from the Philippines where the Department of Agriculture announced plans to seek commercial approval for Vietnamese AVAC ASF vaccines, with the government-run vaccine initiative ongoing. Foot-and-mouth disease restrictions have eased in Germany as no new cases have been reported. If no further outbreaks occur, Germany could regain its FMD-free status after three months, allowing the removal of trade restrictions on meat and dairy products. New Japanese encephalitis virus outbreaks have been confirmed in Australian piggeries, the first since July 2022 in South Queensland. And in the United Kingdom, German imports of meat and dairy products entered the country despite a ban due to the foot-and-mouth disease outbreak in Germany.

PRRS Cumulative Incidence for MSHMP Beginning July 1, 2009

PEDV Cumulative Incidence for MSHMP Beginning May 1, 2013
Copyright 2025 | Swinehealth.org | Website by Heartland Marketing Group